Subramanian Lab Research
We are interested in how micron-length scale structures that are critical for cellular signaling emerge from the collective activity of nanometer sized proteins. We address this question in the context of microtubule organization for (1) cell division and (2) Hedgehog signal transduction
We primarily use a reconstitution-based approach and ‘reconstruct’ sub-reactions of these cellular pathways in vitro from purified components to decipher the fundamental rules that govern their spatial-temporal organization and function. We employ a wide range of experimental approaches, integrating information from cutting edge single-molecule approaches, high-resolution microscopy, structural methods, and biochemical and cellular read-outs.
Through these studies, our goal is to understand the molecular mechanisms relevant to developmental disorders and cancer biology.
How does a dividing cell find its center?
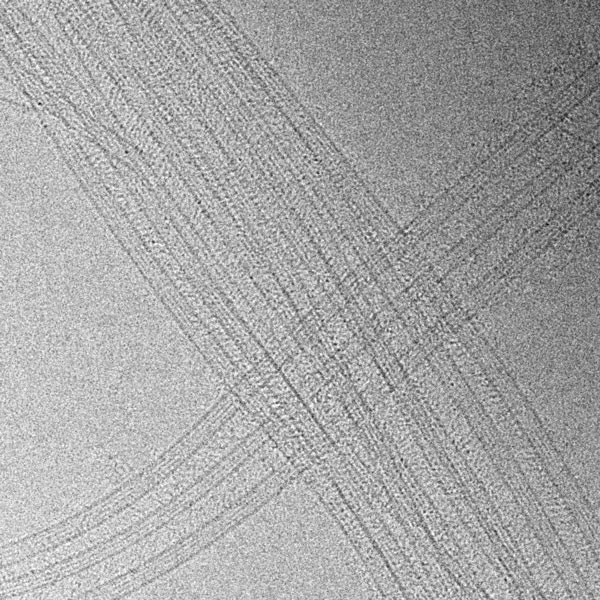
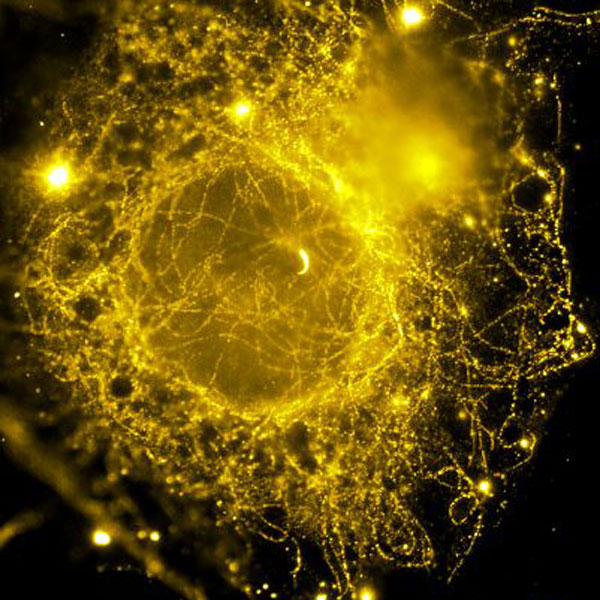
Genome integrity relies on the accurate partitioning of chromosomes during cell division. The spindle midzone is an antiparallel array of microtubules that assembles at the cell center in anaphase and provides the spatial cues that precisely position the cell cleavage furrow. We aim to decipher the mechanisms that govern the organization of the spindle midzone.
Since overlapping microtubule arrays are ubiquitously observed in several cellular contexts such as in neurons and in plant cells, we expect these studies to inform on the fundamental rules that govern the organization of microtubules into well defined arrays for distinct functions.
How is the primary cilium organized for Hedgehog signaling?
The evolutionarily conserved Hedgehog pathway is critical for embryonic development and disrupted in a growing number of tumors. In vertebrates this vital signaling pathway requires the primary cilium, a microtubule-based organelle that acts as a cellular antenna.
It is well established that defects in cilium structure result in aberrant signaling. This surprising finding raises some important questions: How is the primary cilium architecture specialized for Hedgehog signaling? How are signal transduction components spatially localized and precisely regulated within the cilium?
Our long-term goal is to provide insights into these outstanding questions by using two complementary approaches:
(i) in vitro reconstitution of key pathway sub-reactions and
(ii) quantitative live cell imaging of pathway components.
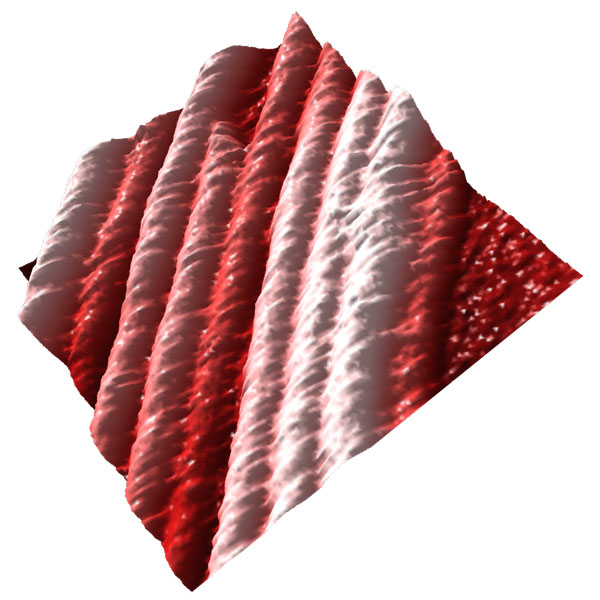
Imaging the dynamics of complex microtubule arrays by Atomic Force Microscopy (AFM)
In this new research direction, we aim to directly visualize the dynamics of complex micron-scale microtubule arrays with single microtubule and protofilament resolution.
By adapting AFM to bridge the spatial-temporal resolution gap between light and electron microscopy, we hope to gain new insights into the mechanisms that assemble and disassemble complex cellular microtubule arrays.